Goals
- Identify spatial clusters with significantly low immunization rates
- Develop an agent-based model for the spread of measles that incorporates detailed immunization data, and is calibrated using a novel source of incidence data
- Develop methods to find and characterize critical spatial clusters, with respect to different metrics, which capture both epidemic and economic burden, and order underimmunized clusters based on their criticality
- Use the methodology to evaluate interventions in terms of their effect on criticality
One fundamental task in network analysis is detecting “hotspots” or “anomalies” in the network; that is, detecting subgraphs where there is significantly more activity than one would expect given historical data or some baseline process. Scan statistics is one popular approach used for anomalous subgraph detection. This methodology involves maximizing a score function over all connected subgraphs, which is a challenging computational problem. A number of heuristics have been proposed for these problems, but they do not provide any quality guarantees. Here, we provide a framework for designing algorithms for optimizing a large class of scan statistics for networks, subject to connectivity constraints. Our algorithms run in time that scales linearly on the size of the graph and depends on a parameter we call the “effective solution size,” while providing rigorous approximation guarantees. In contrast, most prior methods have super-linear running times in terms of graph size. Extensive empirical evidence demonstrates the effectiveness and efficiency of our proposed algorithms in comparison with state-of-the-art methods. Our approach improves on the performance relative to all prior methods, giving up to over 25% increase in the score. Further, our algorithms scale to networks with up to a million nodes, which is 1--2 orders of magnitude larger than all prior applications.
Emergence of under-immunized clusters is a growing concern for public health agencies because they can act as reservoirs of infection and increase the risk of infection into the wider population. We use realistic population models for Minnesota and Washington state, and combine this with school level immunization data to estimate vaccine coverage at the level of census block groups. A scan statistic method defined on networks is used for finding significant clusters of under-immunized block groups, without any restrictions on shape. Further we provide the demographic characteristics of these clusters. We find significant under-vaccinated clusters in MN and WA. These are very irregular in shape, in contrast to the circular disks reported in prior work, which rely on the SatScan approach. Some of the clusters found by our method are not contained in those computed using SatScan, a state-of-the-art software tool used in similar studies in other states. Higher resolution clusters computed using our network based approach and population models provide new insights on the structure and characteristics of such clusters and enable targeted interventions (Cadena et al. BMC Medical Informatics and Decision-Making).
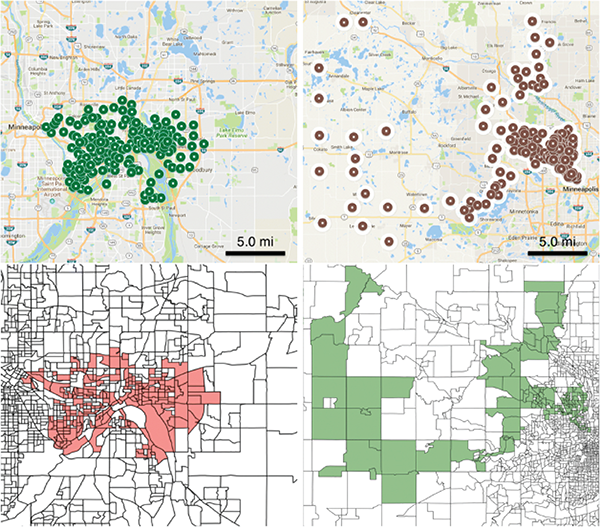
Figure 1: Top two significant clusters in MN (top right and top left) are shown. Each dot on the map is a block group. The same clusters are shown as block group polygons on the bottom right and left, with each marker corresponding to a block group. First cluster in Minnesota covers the city of St. Paul (top left and bottom left) and the second cluster covers the rural block groups west of Minneapolis (top right and bottom right)
Finding Critical Clusters
In this research, we formalize the outbreak risk of a cluster by its “criticality", which is defined as the “probability of a large outbreak" caused by a single case of measles in the cluster. We focus on clusters that are most significant in terms of under immunization rates and measure their criticality. Finding significant under immunized clusters and computing their criticality is a challenging computational problem. We use a synthetic social contact network model for Virginia, and school-level immunization data in the state, along with a network scan statistics approach to find significant under immunized clusters. We combine this with a detailed stochastic agent-based simulation framework to estimate the criticality of each significant cluster, by simulating outbreaks that originate in these clusters (Fig. 2). We also examine how the criticality of clusters changes under a hypothetical 5% drop in MMR rate among children (younger than 12 years), possibly due to disruptions caused by COVID- 19. Finally, we study the demographic, geographic, and network factors associated with such clusters, which can help explain the potential risk of a cluster. Identifying critical clusters and the factors associated with it will be of use to public health authorities who can use this information to prioritize mitigation efforts. Note that although this work focuses on measles, as it uses MMR coverage data and a measles specific disease model, the methodology is generic and can be applied to study other infectious diseases.
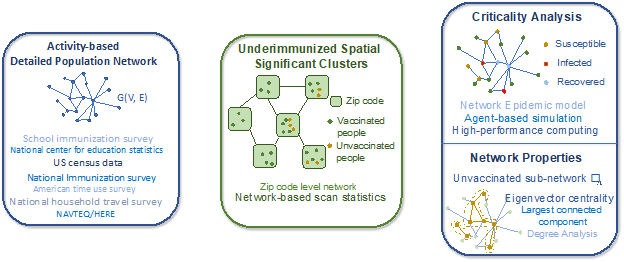
Figure 2. Four major components of the framework: 1) An activity-based population network G(V,E), where a node represents an individual, and an edge represents a contact between two people; 2) under-immunized spatial clusters in a zip code level network Gz(Vz,Ez), where nodes are zip codes, and a connection between two zip codes represents a geographically shared boundary; 3) Criticality analysis of each cluster using the stochastic network epidemic model; and 4) Understanding criticality by investigating network properties of unvaccinated subnetwork Gu. To find significant clusters and their criticality, we first identify statistically significant under immunized clusters; for this purpose, we develop a zip code level spatial network from a detailed activity-based population contact network and use a network scan statistics method. Next, we investigate the criticality of each cluster by importing a single case of measles and simulating its spread using an agent-based model.
Criticality of a cluster. We define the criticality of the set Vc ⊂V in immunization v, denoted by crit(Vc, v), as the probability of a large outbreak, when the initial infection occurs in subset Vc. Formally, crit(Vc, v) = Pr[I(v, Vc)> OC], where OC is the threshold for a large outbreak (taken to be 500, as mentioned above). We use the probability of an outbreak instead of the more commonly used metric of expected number of infections because the expectation is quite small, as many outbreaks die out due to the high immunization rates.
To understand the potential reasons behind the criticality of clusters, we investigate their characteristics such as geographic and network properties. We explore the impact of geographic location, size, population density, and under-immunization rate on the criticality of a cluster. For network attributes, we measure their degree, node strength, and eigenvector centrality. We also investigate the connected components.
Our analysis shows that the local network properties of a cluster are not enough in measuring its potential risk of an outbreak from a drop in immunization rate. We have considered five local properties: size, degree, node strength, links inside of a cluster, and connections inside to outside. A small drop in immunization coverage (only 0.8%) increases the outbreak risk of one of the clusters many folds; however, surprisingly, the increment in local properties of this cluster is the lowest among all the clusters. We find that while the local properties can be misleading, global network properties like the eigenvector analysis can provide a better understanding of the change in criticality, since it considers the full network connectivity. This research shows that the outbreak risk of under-immunized clusters is vastly different depending on their location, size, immunization rate, and network properties.
For more details on this analysis, please see: S Moon*, A Marathe, A Vullikanti. Are all underimmunized measles clusters equally critical? Royal Society Open Science, August 2023. https://doi.org/10.1098/rsos.230873
Other Team Members
Travis Porco | Professor of Global Health | University of California, San Francisco
Nicola Klein | Senior Research Scientist | Kaiser Permanente
Controlling Epidemic Spread using Probabilistic Diffusion Models on Networks
Deploying Vaccine Distribution Sites for Improved Accessibility and Equity to Support Pandemic Response
Deploying Vaccine Distribution Sites for Improved Accessibility and Equity to Support Pandemic Response
Differentially Private Community Detection for Stochastic Block Models
Efficiently Learning the Topology and Behavior of a Networked Dynamical System Via Active Queries
Fair Disaster Containment via Graph-Cut Problems
Fundamental limitations on efficiently forecasting certain epidemic measures in network models
Theoretical Models and Preliminary Results for Contact Tracing and Isolation
Differentially Private Densest Subgraph Detection
Drivers and Predictors of COVID-19 Vaccine Hesitancy in Virginia
Persistence of anti-vaccine sentiment in social networks through strategic interactions
Prioritizing allocation of COVID-19 vaccines based on social contacts increases vaccination effectiveness
Finding Spatial Clusters Susceptible to Epidemic Outbreaks due to Undervaccination
Near-Optimal and Practical Algorithms for Graph Scan Statistics with Connectivity Constraints
Near-Optimal and Practical Algorithms for Graph Scan Statistics
Other Publications
-
S Moon*, A Marathe, A Vullikanti. Are all underimmunized measles clusters equally critical? Royal Society Open Science, August 2023. https://doi.org/10.1098/rsos.230873
-
S Chu*, X Liu*, X Deng, A Marathe. A Latent Process Approach to Change-Point Detection of Mixed-Type Observations. Quality Engineering, 2023, pages 1-20.
-
Truelove, S., Smith, C.P., Qin, M., Mullany, L.C., Borchering, R.K., Lessler, J., Shea, K., Howerton, E., Contamin, L., Levander, J. and Kerr, J., et al. 2022. Projected resurgence of COVID-19 in the United States in July—December 2021 resulting from the increased transmissibility of the Delta variant and faltering vaccination. Elife, 11, p.e73584. PMID: 35726851
-
M Thakur*, R Zhou*, M Mohan*, A Marathe, J Chen, S Hoops, D Machi, B Lewis, A Vullikanti. COVID’s collateral damage: Likelihood of measles resurgence in the United States. BMC Infectious Diseases, September 2022. PMID: 36127637
-
Zirou Qiu*, Baltazar Espinoza*, Vitor V. Vasconcelos, Chen Chen*, Sara Constantino, Stefani Crabtree, Luojun Yang, Anil Vullikanti, Jiangzhuo Chen, Jörgen Weibull, Kaushik Basu, Avinash Dixit, Simon Levin, Madhav Marathe. Understanding the coevolution of mask wearing and epidemics: A network perspective. Proceedings of the National Academy of Sciences. 2022 June; 119(26). PMID: 35733262
-
Borchering RK, Mullany LC, Howerton E, Chinazzi M, Smith CP, Qin M, Reich NG, Contamin L, Levander J, Kerr J, Espino J, Hochheiser H, Lovett K, Kinsey M, Tallaksen K, Wilson S, Shin L, Lemaitre JC, Hulse JD, Kaminsky J, Lee EC, Hill AL, Davis JT, Mu K, Xiong X, Pastore Y Piontti A, Vespignani A, Srivastava A, Porebski P, Venkatramanan S, Adiga A, Lewis B, Klahn B, Outten J, Hurt B, Chen J, Mortveit H, Wilson A, Marathe M, Hoops S, Bhattacharya P, Machi D, Chen S, Paul R, Janies D, Thill JC, Galanti M, Yamana T, Pei S, Shaman J, España G, Cavany S, Moore S, Perkins A, Healy JM, Slayton RB, Johansson MA, Biggerstaff M, Shea K, Truelove SA, Runge MC, Viboud C, Lessler J. Impact of SARS-CoV-2 vaccination of children ages 5-11 years on COVID-19 disease burden and resilience to new variants in the United States, November 2021-March 2022: A multi-model study. Lancet Reg Health Am. 2023 Jan;17:100398. doi: 10.1016/j.lana.2022.100398. Epub 2022 Nov 22. PubMed PMID: 36437905; PubMed Central PMCID: PMC9679449.
-
Venkatramanan S, Sadilek A, Fadikar* A, Barrett CL, Biggerstaff M, Chen J, Dotiwalla X, Eastham P, Gipson B, Higdon D, Kucuktunc O, Lieber A, Lewis BL, Reynolds Z, Vullikanti AK, Wang L, Marathe M. Forecasting influenza activity using machine-learned mobility map.
-
A Basak*, J Cadena*, A Marathe and A Vullikanti. Detection of Spatio-temporal Clusters of Opioid Users with Network Scan Statistics: A Multi-state Analysis. JMIR Public Health and Surveillance, 2019;5(2):e12110 DOI: 10.2196/12110 PMID: 31210142
-
Harrison G, Alabsi Aljundi A, Chen J, Ravi S, Vullikanti A, Marathe M, Adiga A. Identifying Complicated Contagion Scenarios from Cascade Data. Proceedings of the 29th ACM SIGKDD Conference on Knowledge Discovery and Data Mining. KDD '23: 2023; Long Beach CA USA. New York, NY, USA.
-
Wang, L., Adiga, A., Chen, J., Lewis, B., Sadilek, A., Venkatramanan, S. and Marathe, M., 2022. Combining Theory and Data-Driven Approaches for Epidemic Forecasts. Knowledge Guided Machine Learning (pp. 55-82). Chapman and Hall/CRC.